Faculty Profile
 Tasneem Bawa-Khalfe
Tasneem Bawa-Khalfe
Associate Professor
Department of Biology and Biochemistry
Research Division: Biochemistry (Primary)
Office: Science & Engineering Research Center, 3010
Contact: tbawa-khalfe@uh.edu - (713) 743-4288
Education: Ph.D., University of Houston
Google Scholar Profile
Website
The research focus of the Bawa-Khalfe laboratory is on how cancer cells exploit select post-translational modification (PTM) systems to drive cancer progression.
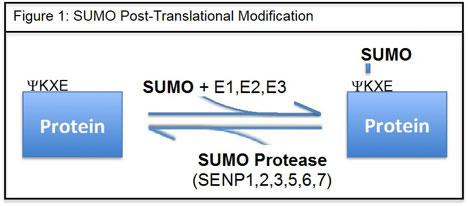
PTMs ensure the structural and functional diversity of a cell’s proteome. Most PTMs are dynamic and closely guarded via the activity of specific enzymes that conjugate or de-conjugate protein targets. Addition of the small ubiquitin-like modifier (SUMO) to a protein is a type of PTM that occurs through a defined sequential process called SUMOylation. SUMOylation includes a SUMO E1 activating complex, the E2 conjugating enzyme Ubc9, and several E3 ligases (Fig. 1). Inversely, the isopeptidase activity of the SUMO proteases (SENP) reverts the substrate to an unmodified state or promotes deSUMOylation.
A large repertoire of cellular proteins is subject to SUMO PTM. SUMOylation affects the conjugated protein’s function, subcellular localization, and/or stability. Consistently, maintaining SUMO dynamics is important for normal cell physiology.
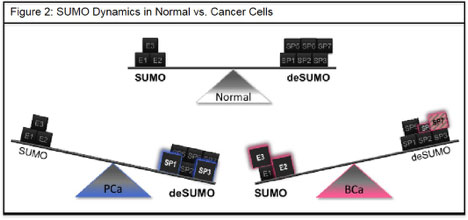
The onset of disease including cancer can alter the transcription, translation, and/or PTM of select components of the SUMO machinery. Naturally, this disturbs the balance of SUMO modified to unmodified substrates (Fig. 2). An imbalance in the kinetics of SUMO PTM can initiate cancer development; for example, induction of SENP1 (SP1) elicits cancerous transformation of the normal mouse prostate gland (Bawa-Khalfe et. al. 2007). Recently, we reported alternative splicing of the SENP7 (SP7) gene product that is readily observed in human metastatic breast cancer samples (Bawa-Khalfe et. al. 2012). However, it is unclear how the loss of SUMO equilibrium supports the cancer transcriptome and an aggressive metastatic phenotype in breast cancer.
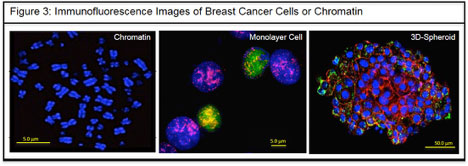
The Bawa-Khalfe Lab is currently applying multiple genomic, epigenetic, and proteomic techniques to address this question; these techniques include chromatin immunoprecipitation (ChIP), ChIP-Seq, RNA-Seq, RNA immunoprecipitation (RIP), mass spectrometry, and in-vitro PTM analysis. The studies are performed at the level of the chromatin, monolayer cell cultures, 3D-acini tumor models (Fig. 3), and genetically engineered whole animal models. Hence, the laboratory’s research integrates results from a single cancer cell and more complex heterogeneous tumor microenvironments. This multipronged approach will provide a more accurate assessment of the therapeutic and/or diagnostic potential of SUMO and other PTM systems in aggressive cancers.