Faculty Profile
 Masaya Fujita
Masaya Fujita
Professor
Department of Biology and Biochemistry
Research Division: Biochemistry (Primary), Cell and Molecular Biology (Joint/Adjunct)
Office: Houston Science Center, 430
Contact: mfujita@uh.edu - (713) 743-9479
Education: Ph.D., Osaka University
Google Scholar Profile
Website
Bacterial differentiation and signal transduction
Dr. Masaya Fujita’s research interests focus on determining how asymmetry is generated during cell development (Fig. 1) and how gene expression is regulated both temporally and spatially (Fig. 2). Bacillus subtilis, a non-pathogenic Gram-positive soil bacterium, is an ideal system in which to explore these fundamental biological questions, as it combines genetic tractability with a variety of cell differentiation programs including sporulation (the transition to an environmentally resistant and metabolically dormant state) and the formation of biofilms (robust multicellular communities embedded in an extracellular matrix).
These differentiation programs are controlled by the master transcriptional regulator, Spo0A, which is activated upon starvation. KinA is the major kinase responsible for the phosphorylation (i.e., activation) of Spo0A. In the widely accepted “Signal” model, it is proposed that KinA is autophosphorylated (i.e., activated) when its amino-terminal “sensor” domain receives an as-yet unknown starvation signal(s), analogous to the activation mechanisms of eukaryotic receptor kinases.
However, the specific mechanisms of KinA activation and the subsequent activation of Spo0A remain elusive because the molecular identity of the hypothetical starvation signal(s) has yet to be clarified.
Fujita aims to address these questions through a synergistic combination of systems and synthetic biology tools, including network perturbations and rewiring, single-cell imaging, and mathematical modeling.
The lab’s recent data indicate that when a threshold concentration of KinA is reached in the cell, the level of active Spo0A becomes sufficient for entry into the various differentiation programs. Thus, Fujita has proposed a new model in which a threshold level of the major kinase KinA acts as a molecular switch to determine entry into sporulation (Threshold model). The lab will attempt to provide further evidence to confirm the “Threshold” model and refute the previous “Signal” model.
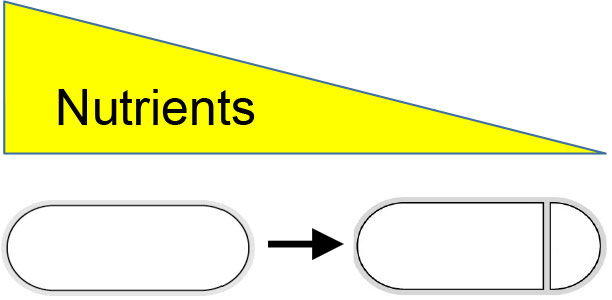
Fig. 1 Starvation induces asymmetric cell division in B. subtilis.
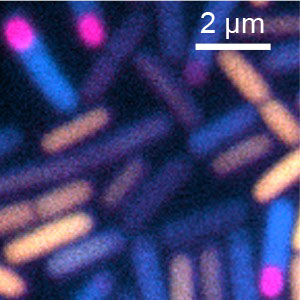
Fig. 2. Stage- and subcellular-specific gene expression in sporulating B. subtilis under starvation. Yellow: growth-specific gene, Cyan: sporulation gene, Magenta: spore-specific gene.
Organizations, Outreach, Boards, Memberships
Member, American Society for Microbiology