GLYCOGEN METABOLISM
- Glycogen = Polymer of glucose with α (1à 4) linkage for the linear chain and, for every 8-14 residues, α (1 à 6) linkage for a branch (Fig. 15-2).
- As 100-400 Å diameter cytoplasmic granules containing up to 120,000 glucose units.
- Highly branched, permits rapid degradation through simultaneous release of glucose units from the end of each branch.
- Liver and muscle are two major storage sites.
GLYCOGEN BREAKDOWN (or Glycogenolysis)
- In Muscle: glycogen → G6P → enetr glycolysis.
- In Liver: glycogen → G6P → G → released into blood.
1. Glycogen Phosphorylase
- (Glu)n + Pi → G1P + (Glu)n-1 where (Glu)n = initial glycogen molecule
- For each cycle, the glucose unit that is released must be at least 5 units from a branch point.
- Catalyzes the rate-limiting step in glycogen breakdown.
2. Glycogen Debranching Enzyme (Fig. 15-6)
- Seeks out a shortened branch with only 4 glucose units.
- Transfers the last 3 units to the 4-OH group at the end of another longer branch.
- Hydrolyzes off the α (1 → 6) linkage of the last glucose unit at the branch point, and releases it as a free glucose. About 10% of glycogen degradation products are free Glu.
3. Phosphoglucomutase
- Requires trace amount of Glucose-1,6-bisphosphate for catalysis.
- G-1,6-bisP
+ Enz ⇌ G6P + Enz-P
- Enz-P +
G1P ⇌ Enz + G-1,6-bisP
-----------------------------------------------------------
- Overall = G1P ⇌ G6P
GLYCOGEN SYNTHESIS
1. UDP-Glucose Pyrophosphorylase
- G1P + UTP ⇌ UDP-glucose + PPi ( ΔG°’ ~ 0 kJ• mol-1 )
- Would be readily reversible. Requires inorganic pyrophosphatase to push the reaction to the right.
- PPi
+ H 2O ⇌ 2 Pi (
ΔG°’ = -19.2 kJ• mol- 1)
- This is a rather common strategy by nature.
2. Glycogen Synthase
- UDP-glucose
+ (Glu) n → UDP + (Glu)n+1
- Elongation of each linear chain.
3. Branching Enzyme (Fig. 15-11)
- Seeks out a linear chain with at least 11 units of glucose in length from the branching point.
- Transfers the terminal chain segment with ~ 7 units of glucose to the C6-OH group of a glucose unit in the same or a different chain.
- This acceptor chain must beat least 4-unit long from an existing branching point.
REGULATION
1. Allosteric (Fig. 15-13)
- Allosteric enzyme: (1) has an active site and at least one separate effector site; (2) the binding of an activator or an inhibitor to an effector site results in a conformational change of the enzyme, (3) the conformational change induced by the activator leads to enzyme activation whereas the conformational change induced by the inhibitor leads to enzyme inactivation.
- Glycogen Phosphorylase: Activator:AMP Inhibitor: ATP, G6P, Glu
- Glycogen Synthase: Activator: G6P
- When ATP is in need: Low ATP, Low G6P, High AMP.
- Results in enhanced activity of Glycogen Phosphorylase and inhibition of Glycogen Synthase. Favors glycogen breakdown.
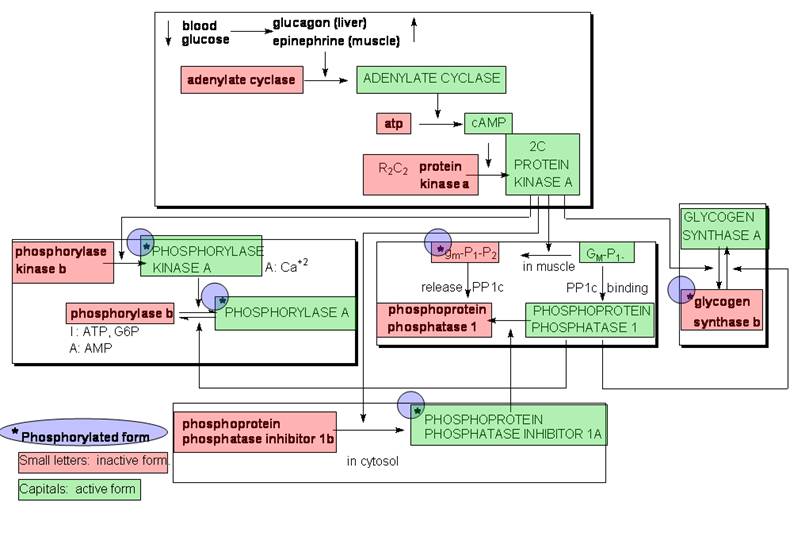
2. Covalent
Modification (Protein Phosphorylation & Dephosphorylation
(A) Phosphorylase a and b
- Phosphorylase a and
b both exist in either the inactive T or the active R conformation (Fig.
15-13). Under physiological conditions, phosphorylase
a is mostly in the R conformation whereas phosphorylase b is mostly T. Hence phosphorylase a is more
active than phosphorylase b.
- Only the T form of phosphorylase
b can be phosphorylated, resulting in the
transformation to phosphorylase a in the T, and in
turn, the R conformation.
(B) Summary of Cascade Regulation (replacing Figs. 15-20, 21)